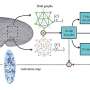
Research at the University of Washington has unveiled a groundbreaking technique known as Deaminase-Assisted single-molecule chromatin Fiber sequencing (DAF-seq). This innovative method significantly enhances the ability to map gene regulation by enabling the analysis of protein interactions along chromatin fibers within single cells at an unprecedented level of detail.
Gene regulation relies heavily on the cooperative binding of proteins to chromatin, the substance that makes up chromosomes. In diploid organisms, which possess two sets of chromosomes, understanding how this protein occupancy varies between different haplotypes—variants of a gene—remains a challenge. The DAF-seq technology addresses this gap, providing insights into the complexities of gene regulation in a way that was previously unattainable.
One of the standout features of DAF-seq is its capability for single-molecule footprinting. This allows researchers to profile chromatin states and DNA sequences with near-nucleotide resolution. The technology generates comprehensive protein occupancy maps that cover approximately 99% of the mappable genome in individual cells. These maps reveal how regulatory elements are occupied by proteins, shedding light on the functional impact of somatic variants and rare chromatin epialleles.
The findings from the DAF-seq approach indicate extensive chromatin plasticity within and between diploid cells. Notably, the research shows that chromatin actuation can diverge by 61% between haplotypes within a single cell, and by 63% across different cells. This significant variation underscores the dynamic nature of gene regulation and the complexity of chromatin behavior.
In addition to revealing variability, DAF-seq also uncovers patterns in how regulatory elements co-actuate along the same chromatin fiber. This behavior is shown to be distance-dependent, reflecting the mechanisms of cohesin-mediated loops, which play a crucial role in organizing chromatin structure and influencing gene expression.
The implications of these discoveries are profound, as they enhance the understanding of gene regulation at a cellular level, potentially impacting fields such as genetics, developmental biology, and medicine. The research was led by a team that includes A.B. Stergachis and others at the University of Washington, with support from various funding organizations including the National Institutes of Health and the Chan Zuckerberg Initiative.
The work has been recognized for its innovative approach and its potential to transform how scientists study gene regulation. As the field moves forward, DAF-seq could become an essential tool for researchers seeking to unravel the complexities of gene interactions and the implications for human health and disease.
This pioneering research not only advances scientific understanding but also opens doors for future studies that could lead to novel therapeutic strategies. For those interested in the intricate world of gene regulation, following developments stemming from DAF-seq will be imperative.